- 多(2)次元正規分布の混成でできているような分布が2つある
- 分布は場所によって、片方の分布から発生しやすかったり、逆にもう片方の分布から発生しやすかったりする
- それは尤度比
- そんなことを理論値や標本からの推定値でプロットしてみる
library(pracma)
library(MCMCpack)
library(ggplot2)
k <- 2
n <- 8
centers <- matrix(runif(n*k),ncol=k)
sds <- list()
for(i in 1:n){
tmp <- t(rdirichlet(k,rep(1,100)))
sds[[i]] <- cov(tmp)
sds[[i]] <- sds[[i]]/sum(sds[[i]])*0.05
}
ng <- 2
library(mvtnorm)
ratios <- matrix(0, ng,n)
for(i in 1:ng){
ratios[i,] <- rdirichlet(1, rdirichlet(1,rep(1,n)*10))
}
ratios <- rdirichlet(ng, rep(1,n))
x <- list(seq(from = -2, to = 3, length = 100))
X <- expand.grid(rep(x,k))
D <- list()
for(i in 1:n){
D[[i]] <- apply(X, 1, dmvnorm, mean = centers[i,], sigma = sds[[i]])
}
D.g <- list()
for(i in 1:ng){
D.g[[i]] <- rep(0,length(D[[1]]))
for(j in 1:n){
D.g[[i]] <- D.g[[i]] + ratios[i,j] * D[[j]]
}
}
par(mfcol=c(1,2))
image(matrix(D.g[[1]],length(x[[1]]),length(x[[1]])))
image(matrix(D.g[[2]],length(x[[1]]),length(x[[1]])))
par(mfcol=c(1,1))
df1 <- data.frame(x=rep(X[,1],2),y=rep(X[,2],2),z=c(D.g[[1]],D.g[[2]]),gr=factor(c(rep(1,length(X[,1])),rep(2,length(X[,1])))))
v <- ggplot(df1, aes(x, y, z = z))
v +geom_tile(aes(fill=z)) + scale_fill_gradient(low="green", high="red")+ stat_contour() + facet_wrap(~ gr)
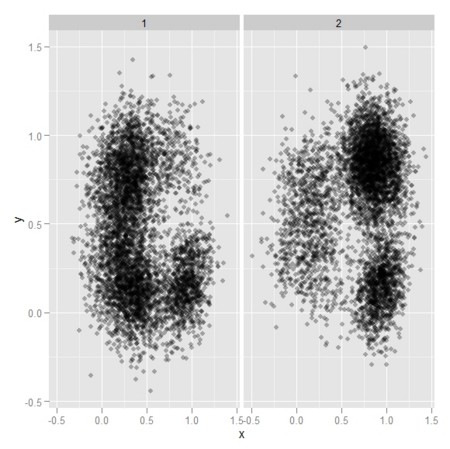
df <- data.frame(x=c(R[[1]][,1],R[[2]][,1]),y=c(R[[1]][,2],R[[2]][,2]),gr = c(rep(1,Npt[1]+n),rep(2,Npt[2]+n)))
gp <- ggplot(df,aes(x=x,y=y,colour = factor(gr))) + geom_point(alpha=0.3)
gp
selected <- sample(1:length(R[[1]][,1]),100)
LR <- rep(0,length(selected))
LR.gauss <- LR
for(i in 1:length(LR)){
LR[i] <- likelihoodratio.roc.kernel(R[[1]],R[[2]],c(R[[1]][selected[i],]),kernel="g")$dens.0
LR.gauss[i] <- likelihoodratio.gauss(R[[1]],R[[2]],c(R[[1]][selected[i],]))$LR
}
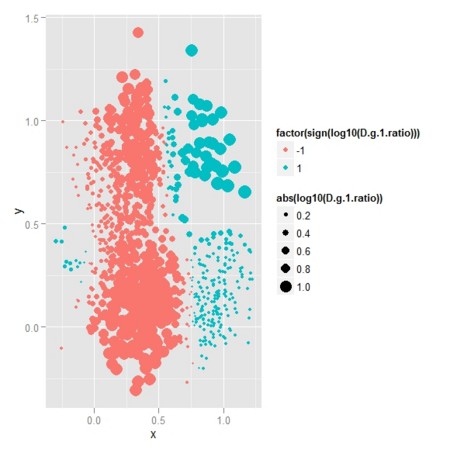
df.new <- data.frame(x=R[[1]][selected,1],y=R[[1]][selected,2],z=LR)
ggg <- ggplot(data = df.new,aes(x=x,y=y,size=abs(log10(z)),colour=factor(sign(log10(z))))) + geom_point()
ggg
df.new.gauss <- data.frame(x=R[[1]][selected,1],y=R[[1]][selected,2],z=LR.gauss)
ggg.gauss <- ggplot(data = df.new.gauss,aes(x=x,y=y,size=abs(log10(z)),colour=factor(sign(log10(z))))) + geom_point()
dev.new()
ggg.gauss
dev.new()
gp <- ggplot(df,aes(x=x,y=y)) + geom_point(alpha=0.3) + facet_wrap(~ gr)
gp
D.1 <- list()
for(i in 1:n){
D.1[[i]] <- apply(R[[1]][selected,], 1, dmvnorm, mean = centers[i,], sigma = sds[[i]])
}
D.g.1 <- list()
for(i in 1:ng){
D.g.1[[i]] <- rep(0,length(D.1[[1]]))
for(j in 1:n){
D.g.1[[i]] <- D.g.1[[i]] + ratios[i,j] * D.1[[j]]
}
}
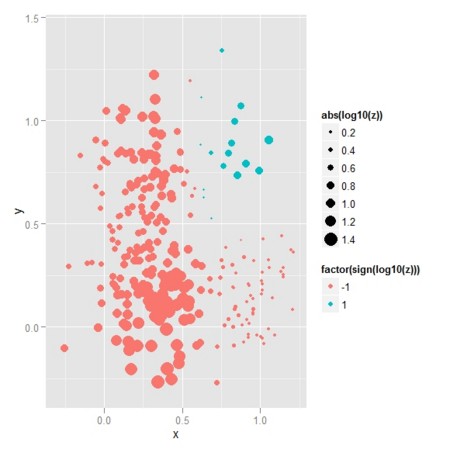
D.g.1.ratio <- D.g.1[[2]]/D.g.1[[1]]
dev.new()
ggg.2 <- ggplot(data = df.new,aes(x=x,y=y,size=abs(log10(D.g.1.ratio)),colour=factor(sign(log10(D.g.1.ratio))))) + geom_point()
ggg.2
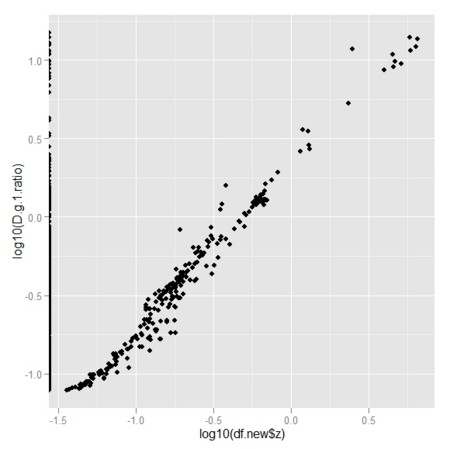
qplot(log10(df.new$z),log10(D.g.1.ratio))
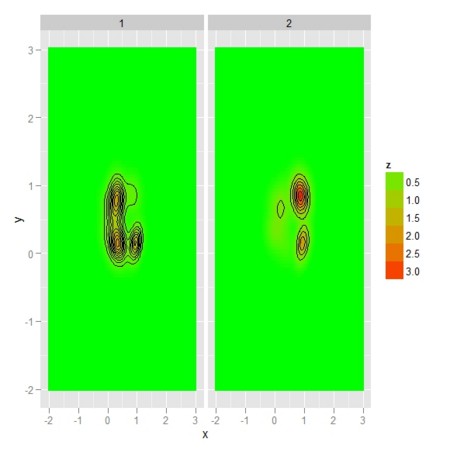
- 理論的尤度比(2色は、どちらの群が優位かを表し、点のサイズは尤度比の差の大きさを表す)
library(sde)
library(pracma)
k <-2
c1 <- rep(0,k)
c2 <- c(1,rep(0,k-1))
x <- y <- seq(from=-2,to=3,by=pi/30)
xy <- expand.grid(x,y)
sd1 <- 0.5
sd2 <- 0.7
Npt <- 10000
A.1 <- matrix(rnorm(Npt*k,sd=sd1),ncol=k)
A.2 <- matrix(rnorm(Npt*k,sd=sd2),ncol=k)
A.2[,1] <- A.2[,1]+c2[1]
my.A <- data.frame(x=c(A.1[,1],A.2[,1]),y=c(A.1[,2],A.2[,2]),group= c(rep(1,length(A.1[,1])),rep(2,length(A.2[,1]))))
gp <- ggplot(data = my.A)
gp <- gp + aes(x=x,y=y,colour=factor(group)) + geom_point(alpha=0.3)
gp
dens.calc <- function(x,t){
y <- t(t(x)-t)
r <- sqrt(apply(y^2,1,sum))
n <- length(x[1,])
s <- Sphere.surface(n-1,r)
return(list(r=r,s=s))
}
Sphere.surface <- function(n, r){
2*pi^((n+1)/2) / gamma((n+1)/2) * r^n
}
my.roc<-function(x,y,t=NULL){
Nx <- length(x)
Ny <- length(y)
if(is.null(t))t<-seq(from=min(x,y),to=max(x,y),length=100)
xa<-outer(x,t,FUN="<")
ya<-outer(y,t,FUN="<")
xb<-apply(sign(xa),2,sum)
yb<-apply(sign(ya),2,sum)
xy<-data.frame(X=Nx-xb,Y=Ny-yb)
return(xy)
}
my.roc.2<-function(x,y,t=NULL){
Nx <- length(x)
Ny <- length(y)
if(is.null(t))t<-seq(from=min(x,y),to=max(x,y),length=100)
xa<-outer(x,t,FUN="<")
ya<-outer(y,t,FUN="<")
xb<-apply(sign(xa),2,sum)
yb<-apply(sign(ya),2,sum)
X<-Nx-xb
Y<-Ny-yb
X.1<-(X-min(X))/(max(X)-min(X)) *2-1
Y.1<-(Y-min(Y))/(max(Y)-min(Y)) *2-1
xy<-data.frame(X=X.1,Y=Y.1)
return(xy)
}
glmPolynomial<-function(y,x,dg){
Xs<-matrix(0,length(x),dg)
for(i in 1:dg){
Xs[,i]<-x^i
}
fit<-glm(y~Xs[,1:dg])
beta<-fit$coefficients
Xs2<-cbind(rep(1,length(x)),Xs)
predY<-t(t(Xs2)*beta)
sumupPredY<-apply(predY,1,sum)
list(beta=beta,x=x,predY=sumupPredY)
}
poly.grad <- function(y,x,dg,t){
glmpoly <- glmPolynomial(y,x,dg)
ret <- rep(0,length(t))
for(i in 1:(dg-1)){
ret <- ret + glmpoly$beta[i+1]*i*t^(i-1)
}
list(glmpoly=glmpoly,deriv=ret)
}
x<-sort(runif(100))
y<-runif(100)+x^3*2+x^4*5
xlim<-c(min(x),max(x))
ylim<-c(min(y),max(y))
plot(x,y,xlim=xlim,ylim=ylim,col="red")
dgs<-1:5
for(i in dgs){
outglm<-glmPolynomial(y,x,i)
par(new=TRUE)
plot(outglm$x,outglm$predY,xlim=xlim,ylim=ylim,col=gray(i/(max(dgs)*1.5)),type="l")
}
tmp.fx <- function(A.1,A.2,P,npt = 512, theta=pi/6,deg=8,deriv.pt=NULL){
d1 <- dens.calc(A.1,P)
d2 <- dens.calc(A.2,P)
R1 <- c(d1$r,-d1$r)
R2 <- c(d2$r,-d2$r)
t.2 <- seq(from=min(R1,R2),to=max(R1,R2),length=npt)
my.roc.out <- my.roc.2(R1,R2,t.2)
out.df <- data.frame(thres=t.2,roc.x=my.roc.out$X,roc.y=my.roc.out$Y)
myroc.df <- data.frame(x=my.roc.out$X,y=my.roc.out$Y)
gg.roc <- ggplot(data=myroc.df,aes(x=x,y=y))
gg.roc <- gg.roc + geom_point() + stat_smooth()
M <- matrix(c(cos(theta),cos(pi/2-theta),sin(theta),sin(pi/2-theta)),byrow=TRUE,2,2)
new.XY<-t(M%*%t(as.matrix(myroc.df)))
out.df$roc.x.rotated=new.XY[,1]
out.df$roc.y.rotated=new.XY[,2]
new.XY.df <- data.frame(x=new.XY[,1],y=new.XY[,2])
n<-length(new.XY.df$x)
x <- out.df$roc.x.rotated
y <- out.df$roc.y.rotated
if(is.null(deriv.pt)){
deriv.pt <- sort(c(x,0))
}
poly.grad.out <- poly.grad(y,x,deg,deriv.pt)
glmpoly <- poly.grad.out$glmpoly
beta<-glmpoly$beta
out.df$approx.x.rotated <- glmpoly$x
out.df$approx.y.rotated <- glmpoly$predY
grad.df <- data.frame(grad.x=deriv.pt,grad.rotated=poly.grad.out$deriv)
df.poly <- data.frame
phi <- atan(grad.df$grad.rotated)
grad.df$phi.rotated <- phi
phi<-(phi-pi/4)*(pi/4)/(pi/4-theta)+pi/4
grad.df$phi <- phi
return(list(data.df = out.df,grad.df=grad.df,poly.beta=beta))
}
likelihoodratio.roc.poly <- function(A.1,A.2,P,npt = 512, theta=pi/6,deg=8,deriv.pt=NULL){
d1 <- dens.calc(A.1,P)
d2 <- dens.calc(A.2,P)
R1 <- c(d1$r,-d1$r)
R2 <- c(d2$r,-d2$r)
t.2 <- seq(from=min(R1,R2),to=max(R1,R2),length=npt)
my.roc.out <- my.roc.2(R1,R2,t.2)
out.df <- data.frame(thres=t.2,roc.x=my.roc.out$X,roc.y=my.roc.out$Y)
myroc.df <- data.frame(x=my.roc.out$X,y=my.roc.out$Y)
M <- matrix(c(cos(theta),cos(pi/2-theta),sin(theta),sin(pi/2-theta)),byrow=TRUE,2,2)
new.XY<-t(M%*%t(as.matrix(myroc.df)))
out.df$roc.x.rotated=new.XY[,1]
out.df$roc.y.rotated=new.XY[,2]
new.XY.df <- data.frame(x=new.XY[,1],y=new.XY[,2])
x <- out.df$roc.x.rotated
y <- out.df$roc.y.rotated
if(is.null(deriv.pt)){
deriv.pt <- sort(c(x,0))
}
poly.grad.out <- poly.grad(y,x,deg,deriv.pt)
glmpoly <- poly.grad.out$glmpoly
beta<-glmpoly$beta
out.df$approx.x.rotated <- glmpoly$x
out.df$approx.y.rotated <- glmpoly$predY
grad.df <- data.frame(grad.x=deriv.pt,grad.rotated=poly.grad.out$deriv)
df.poly <- data.frame
phi <- atan(grad.df$grad.rotated)
grad.df$phi.rotated <- phi
phi<-(phi-pi/4)*(pi/4)/(pi/4-theta)+pi/4
grad.df$phi <- phi
return(list(data.df = out.df,grad.df=grad.df,poly.beta=beta))
}
t.out <- tmp.fx(A.1,A.2,rep(0,k),theta=pi/10,deg=8,npt=10^4,deriv.pt=seq(from=-1,to=1,length=513))
gg.1 <- ggplot(data=t.out$data.df, aes(x=roc.x,y=roc.y)) + geom_point()
gg.1
gg.2 <- ggplot(data=t.out$data.df, aes(x=roc.x.rotated,y=roc.y.rotated)) + geom_point()
gg.2
gg.3 <- ggplot(data=t.out$data.df, aes(x=approx.x.rotated,y=approx.y.rotated)) + geom_point()
gg.3
gg.4 <- ggplot(data=t.out$grad.df, aes(x=grad.x,y=grad.rotated)) + geom_point()
gg.4
gg.5 <- ggplot(data=t.out$grad.df, aes(x=grad.x,y=phi.rotated)) + geom_point()
gg.5
gg.6 <- ggplot(data=t.out$grad.df, aes(x=grad.x,y=phi)) + geom_point()
gg.6
gg.7 <- ggplot(data=t.out$grad.df, aes(x=grad.x,y=tan(phi))) + geom_point()
gg.7
tan(t.out$grad.df$phi[which(t.out$grad.df$grad.x==0)])
likelihoodratio.roc.kernel <- function(A.1,A.2,P,npt.roc = 1000,npt.kernel = 10^6, density.npt = 513,kernel= c("gaussian", "epanechnikov", "rectangular","triangular", "biweight","cosine", "optcosine")){
d1 <- dens.calc(A.1,P)
d2 <- dens.calc(A.2,P)
R1 <- c(d1$r,-d1$r)
R2 <- c(d2$r,-d2$r)
t <- seq(from=min(R1,R2),to=max(R1,R2),length=npt.roc)
my.roc.out <- my.roc.2(R1,R2,t)
x <- my.roc.out$X
y <- my.roc.out$Y
yi<-seq(from=range(y)[1],to=range(y)[2],length=npt.kernel)
xc<-interp1(y,x,yi,method="constant")
density.out <- density(xc,n=density.npt,from=-1,to=1,kernel=kernel)
return(list(roc.df = data.frame(t=t,x=x,y=y),interpor.roc.df = data.frame(x=xc,y=yi),density.out=density.out,dens.0=density.out$y[which(density.out$x==0)]))
}
likelihoodratio.gauss <- function(A.1,A.2,P){
d1 <- dens.calc(A.1,P)
d2 <- dens.calc(A.2,P)
R1 <- d1$r^2
R2 <- d2$r^2
Prob1 <- sum(exp(-R1))/length(R1)
Prob2 <- sum(exp(-R2))/length(R2)
return(list(pr.1 = Prob1, pr.2 = Prob2, LR = Prob1/Prob2))
}




