diffEqパッケージで数値微分
- こちらのPDFをただコピーして実行してみるだけ

Solving Differential Equations in R (Use R!)
- 作者: Karline Soetaert,Jeff Cash,Francesca Mazzia
- 出版社/メーカー: Springer
- 発売日: 2012/06/07
- メディア: ペーパーバック
- この商品を含むブログを見る
- この本の10章の話
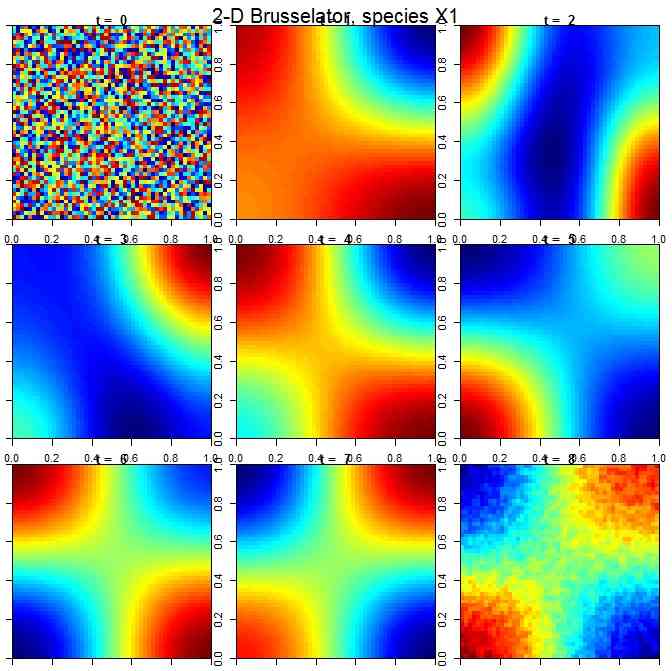
library(diffEq) N <- 100 xgrid <- setup.grid.1D(x.up = 0, x.down = 1, N = N) x <- xgrid$x.mid D.coeff <- 0.01 Diffusion <- function (t, Y, parms){ tran <- tran.1D(C = Y, C.up = 0, C.down = 1, D = D.coeff, dx = xgrid) list(dY = tran$dC, flux.up = tran$flux.up, flux.down = tran$flux.down) } Yini <- sin(pi*x) times <- seq(from = 0, to = 5, by = 0.01) print(system.time( out <- ode.1D(y = Yini, times = times, func = Diffusion, parms = NULL, dimens = N) )) par (mfrow=c(1, 2)) plot(out[1, 2:(N+1)], x, type = "l", lwd = 2,xlab = "Variable, Y", ylab = "Distance, x") for (i in seq(2, length(times), by = 50)) lines(out[i, 2:(N+1)], x) image(out, grid = x, mfrow = NULL, ylab = "Distance, x",main = "Y") dx <- 0.2 xgrid <- setup.grid.1D(x.up = -100, x.down = 100, dx.1 = dx) x <- xgrid$x.mid N <- xgrid$N lam <- 0.05 uini <- exp(-lam*x^2) vini <- rep(0, N) yini <- c(uini, vini) times <- seq (from = 0, to = 50, by = 1) wave <- function (t, y, parms) { u <- y[1:N] v <- y[(N+1):(2*N)] du <- v dv <- tran.1D(C = u, C.up = 0, C.down = 0, D = 1, dx = xgrid)$dC return(list(c(du, dv))) } out <- ode.1D(func = wave, y = yini, times = times, parms = NULL, method = "adams", dimens = N, names = c("u", "v")) u <- subset(out, which = "u") analytic <- function (t, x) 0.5 * (exp(-lam * (x+1*t)^2 ) +exp(-lam * (x-1*t)^2) ) OutAna <- outer(times, x, FUN = analytic) max(abs(u - OutAna)) outtime <- seq(from = 0, to = 50, by = 10) matplot.1D(out, which = "u", subset = time %in% outtime, grid = x, xlab = "x", ylab = "u", type = "l", lwd = 2, xlim = c(-50, 50), col = c("black", rep("darkgrey", 5))) legend("topright", lty = 1:6, lwd = 2, col = c("black", rep("darkgrey", 5)), title = "t = ", legend = outtime) Nx <- 100 Ny <- 100 xgrid <- setup.grid.1D (x.up = 0, x.down = 1, N = Nx) ygrid <- setup.grid.1D (x.up = 0, x.down = 1, N = Ny) x <- xgrid$x.mid y <- ygrid$x.mid laplace <- function(t, U, parms) { w <- matrix(nrow = Nx, ncol = Ny, data = U) dw <- tran.2D(C = w, C.x.up = 0, C.x.down = 0, flux.y.up = 0, flux.y.down = -1 * sin(pi*x)*pi*sinh(pi), D.x = 1, D.y = 1, dx = xgrid, dy = ygrid)$dC list(dw) } print(system.time( out <- steady.2D(y = runif(Nx*Ny), func = laplace, parms = NULL, nspec = 1, dimens = c(Nx, Ny), lrw = 1e7) )) w <- matrix(nrow = Nx, ncol = Ny, data = out$y) analytic <- function (x, y) sin(pi*x) * cosh(pi*y) OutAna <- outer(x, y, FUN = analytic) max(abs(w - OutAna)) image(out, grid = list(x, y), main = "elliptic Laplace", add.contour = TRUE) adv.func <- function(t, y, p, adv.method) list(advection.1D(C = y, C.up = y[N], C.down = y[1], v = 0.1, adv.method = adv.method, dx = xgrid)$dC) xgrid <- setup.grid.1D(0.3, 1.3, N = 50) x <- xgrid$x.mid N <- length(x) yini <- sin(pi * x)^50 times <- seq(0, 20, 0.01) out1 <- ode.1D(y = yini, func = adv.func, times = times, parms = NULL, method = "euler", dimens = N, adv.method = "muscl") out2 <- ode.1D(y = yini, func = adv.func, times = times, parms = NULL, method = "euler", dimens = N, adv.method = "super") N <- 50 Grid <- setup.grid.1D(x.up = 0, x.down = 1, N = N) x1ini <- 1 + sin(2 * pi * Grid$x.mid) x2ini <- rep(x = 3, times = N) yini <- c(x1ini, x2ini) brusselator1D <- function(t, y, parms) { X1 <- y[1:N] X2 <- y[(N+1):(2*N)] dX1 <- 1 + X1^2*X2 - 4*X1 + tran.1D (C = X1, C.up = 1, C.down = 1, D = 0.02, dx = Grid)$dC dX2 <- 3*X1 - X1^2*X2 + tran.1D (C = X2, C.up = 3, C.down = 3, D = 0.02, dx = Grid)$dC list(c(dX1, dX2)) } times <- seq(from = 0, to = 10, by = 0.1) print(system.time( out <- ode.1D(y = yini, func = brusselator1D, times = times, parms = NULL, nspec = 2, names = c("X1", "X2"), dimens = N) )) par(mfrow = c(2, 2)) image(out, mfrow = NULL, grid = Grid$x.mid, which = "X1", method = "contour") image(out, mfrow = NULL, grid = Grid$x.mid, which = "X1") par(mar = c(1, 1, 1, 1)) image(out, mfrow = NULL, grid = Grid$x.mid, which = "X1", method = "persp", col = NA) image(out, mfrow = NULL, grid = Grid$x.mid, which = "X1", method = "persp", border = NA, shade = 0.3 ) brusselator2D <- function(t, y, parms) { X1 <- matrix(nrow = Nx, ncol = Ny, data = y[1:(Nx*Ny)]) X2 <- matrix(nrow = Nx, ncol = Ny, data = y[(Nx*Ny+1) : (2*Nx*Ny)]) dX1 <- 1 + X1^2*X2 - 4*X1 + tran.2D (C = X1, D.x = D_X1, D.y = D_X1, dx = Gridx, dy = Gridy)$dC dX2 <- 3*X1 - X1^2*X2 + tran.2D (C = X2, D.x = D_X2, D.y = D_X2, dx = Gridx, dy = Gridy)$dC list(c(dX1, dX2)) } Nx <- 50 Ny <- 50 Gridx <- setup.grid.1D(x.up = 0, x.down = 1, N = Nx) Gridy <- setup.grid.1D(x.up = 0, x.down = 1, N = Ny) D_X1 <- 2 D_X2 <- 8*D_X1 X1ini <- matrix(nrow = Nx, ncol = Ny, data = runif(Nx*Ny)) X2ini <- matrix(nrow = Nx, ncol = Ny, data = runif(Nx*Ny)) yini <- c(X1ini, X2ini) times <- 0:8 print(system.time( out <- ode.2D(y = yini, parms = NULL, func = brusselator2D, nspec = 2, dimens = c(Nx, Ny), times = times, lrw = 2000000, names=c("X1", "X2")) )) par(oma = c(0,0,1,0)) image(out, which = "X1", xlab = "x", ylab = "y", mfrow = c(3, 3), ask = FALSE, main = paste("t = ", times), grid = list(x = Gridx$x.mid, y = Gridy$x.mid)) mtext(side = 3, outer = TRUE, cex = 1.25, line = -1, "2-D Brusselator, species X1") Nr <- 100 Np <- 100 r <- seq(2, 4, len = Nr+1) theta <- seq(0, 2*pi, len = Np+1) theta.mid <- 0.5*(theta[-1] + theta[-Np]) Model <- function(t, C, p) { y = matrix(nrow = Nr, ncol = Np, data = C) tran <- tran.polar (y, D.r = 1, r = r, theta = theta, C.r.up = 0, C.r.down = 4 * sin(5*theta.mid), cyclicBnd = 2) list(tran$dC) } STD <- steady.2D(y = runif(Nr*Np), parms = NULL, func = Model, dimens = c(Nr, Np), lrw = 1e6, cyclicBnd = 2) OUT <- polar2cart (STD, r = r, theta = theta, x = seq(-4, 4, len = 400), y = seq(-4, 4, len = 400)) image(OUT, main = "Laplace") Nx <- 80 Ny <- 80 xgrid <- setup.grid.1D(-7, 7, N=Nx) ygrid <- setup.grid.1D(-7, 7, N=Ny) x <- xgrid$x.mid y <- ygrid$x.mid sinegordon2D <- function(t, C, parms) { u <- matrix(nrow = Nx, ncol = Ny, data = C[1 : (Nx*Ny)]) v <- matrix(nrow = Nx, ncol = Ny, data = C[(Nx*Ny+1) : (2*Nx*Ny)]) dv <- tran.2D (C = u, C.x.up = 0, C.x.down = 0, C.y.up = 0, C.y.down = 0, D.x = 1, D.y = 1, dx = xgrid, dy = ygrid)$dC - sin(u) list(c(v, dv)) } peak <- function (x, y, x0 = 0, y0 = 0) exp(-((x-x0)^2 + (y-y0)^2)) uini <- outer(x, y, FUN = function(x, y) peak(x, y, 2,2) + peak(x, y,-2,-2) + peak(x, y,-2,2) + peak(x, y, 2,-2)) vini <- rep(0, Nx*Ny) times <- 0:3 print(system.time( out <- ode.2D (y = c(uini, vini), times = times, parms = NULL, func = sinegordon2D, names = c("u", "v"), dimens = c(Nx, Ny), method = "ode45") )) mr <- par(mar = c(0, 0, 1, 0)) image(out, main = paste("time =", times), which = "u", grid = list(x = x, y = y), method = "persp", border = NA, col = "grey", box = FALSE, shade = 0.5, theta = 30, phi = 60, mfrow = c(2, 2), ask = FALSE) par(mar = mr) alf <- 0.5 gam <- 1 Schrodinger <- function(t, u, parms) { du <- 1i * tran.1D (C = u, D = 1, dx = xgrid)$dC + 1i * gam * abs(u)^2 * u list(du) } N <- 300 xgrid <- setup.grid.1D(-20, 80, N = N) x <- xgrid$x.mid c1 <- 1 c2 <- 0.1 sech <- function(x) 2/(exp(x) + exp(-x)) soliton <- function (x, c1) sqrt(2*alf/gam) * exp(0.5*1i*c1*x) * sech(sqrt(alf)*x) yini <- soliton(x, c1) + soliton(x-25, c2) times <- seq(0, 40, by = 0.1) print(system.time( out <- ode.1D(y = yini, parms = NULL, func = Schrodinger, times = times, dimens = 300, method = "adams") )) user system elapsed 2.18 0.03 2.28 image(abs(out), grid = x, ylab = "x", main = "two solitons")